On Aug 10th, 2023, Shixian Lin’s Lab from Life Sciences Institute, Zhejiang University, published a research article in Nature Chemical Biology entitled “Computational design and genetic incorporation of lipidation mimics in living cells”.
Post-translational modification of proteins with lipids, the most hydrophobic moieties in the cell, reshapes protein-membrane and protein-protein interactions, and thereby, dramatically affects protein structure, localization, and trafficking. Given its extreme hydrophobicity, protein lipidation conceivably plays a central role in almost all membrane associated biological pathways, such as cell signaling, protein secretion, cell death and immunity. In this context, eukaryotic cells have evolved dozens of enzymes and a range of regulatory proteins to ensure the correct addition and removal of lipid modifications at the desired sites for hundreds of substrate proteins. However, since lipidation is highly variable (long, medium, and short lipidations at the same site may display very different cellular functions), reversible and often cross-talks with other protein post-translational modifications (PTMs), understanding the biological and physiological functions of lipidation remains challenging; and the common loss-of-function mutagenesis approaches are often not well suited for exploring protein lipidation. Furthermore, lipidation is a clinically-proven PTM that has shown great promise in a variety of top-selling drugs. The generation of lipidated recombinant polypeptides was typically achieved by direct chemical modification of the polypeptide, an approach highly challenging for most proteins due to the poor solubility of hydrophobic lipid mimic that requires solvation in an organic solvent. Although site-specific lipidation has succeeded in a few peptides for therapeutic purposes, most protein drugs could not survive through organic labeling conditions. Co-translational incorporation of lipid mimics into protein in a site-specific manner could efficiently circumvent these limitations.
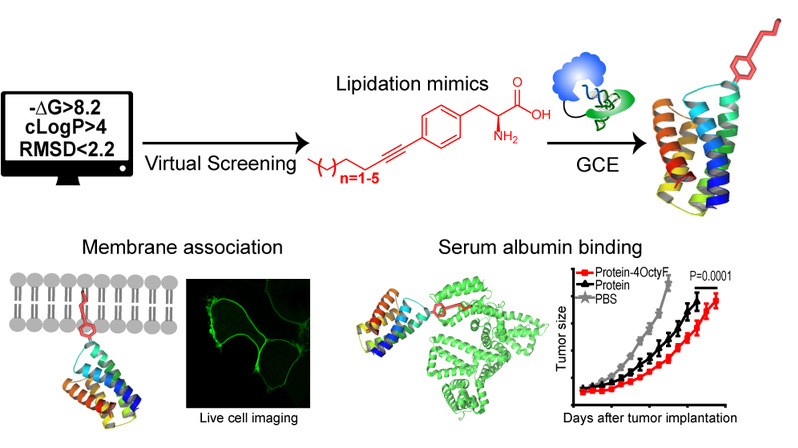
Figure 1. Computational design and genetic incorporation of lipidation mimics in living cells for biological application and therapeutic advance.
Herein, the authors report a computational approach for designing lipidation mimics that fully recapitulate the biochemical properties of natural lipidation in membrane association and albumin binding. Further, they establish an engineered system for co-translational incorporation of these lipidation mimics into any desired position of proteins in E. coli and mammalian cells. they demonstrate the utility of these length-tunable lipidation mimics in diverse applications, including improving the half-life and activity of therapeutic proteins in living mice, anchoring functional proteins to membrane by substituting natural lipidation, functional characterization of protein carrying different lengths of lipidation, and determining the plasma membrane binding capacity of a given compound. The study enables gain-of-function studies of lipidation in hundreds of proteins and facilitates the creation of superior therapeutic candidates.
Dr. Wenlong Ding and Chao Liu are the co-first authors and Dr. Shixian Lin is the corresponding author in this study. This research was supported by the National Key R&D Program of China (Grant 2019YFA09006600), the National Natural Science Foundation of China (Grants 22222705, 92253302, 91953113, 21877096, and 22207095), and China Postdoctoral Science Foundation (grant 2019M652072).
Link: https://www.nature.com/articles/s41589-023-01400-8



