On April 5, 2022, Shixian Lin’s Lab in our institute published a research article in JACS entitled “Manipulating cation-π interactions with genetically encoded tryptophan derivatives”.
Cation-π interactions are the major non-covalent interactions for molecular recognition and play a central role in a broad area of chemistry and biology. Despite tremendous success in understanding the origin and biological importance of cation-π interactions, the design and synthesis of stronger cation-π interactions remains elusive. In this paper, the authors report an approach that greatly increases the binding energy of cation-π interactions by replacing Trp in the aromatic box with an electron-rich Trp derivative using the genetic code expansion strategy. The binding affinity between histone H3K4me3 and its reader is increased by more than 8-fold using genetically encoded 6-methoxy-Trp. Furthermore, through a systematic engineering process, the authors construct an H3K4me3 Super-Reader with single-digit nM affinity for H3K4me3 detection and imaging. More broadly, this approach paves the way for manipulating cation-π interactions for a variety of applications.
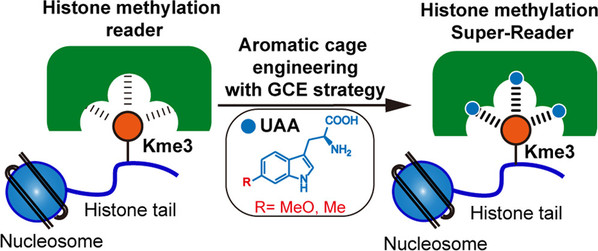
Figure 1. Tuning cation-π interactions with genetic code expansion strategy.
Dr. Hongxia Zhao and Chao Liu are co-first authors in this study. Dr. Shixian Lin is the corresponding author. This research was supported by the National Key R&D Program of China (Grant 2019YFA09006600), the National Natural Science Foundation of China (Grant 91953113, 21877096), and the Fundamental Research Funds for the Zhejiang Provincial Universities (Grant 2019XZZX003-19).
Link:https://pubs.acs.org/doi/10.1021/jacs.1c12944



